WGCNA for male lncRNAs added to WGCNA code for females. Link to code.
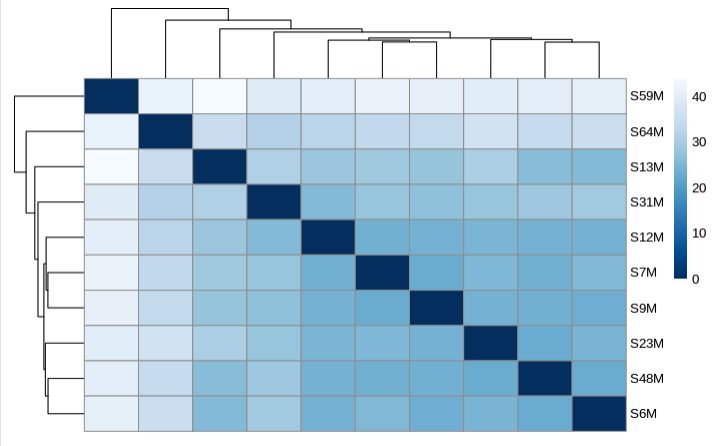
Heatmap of sample distances:
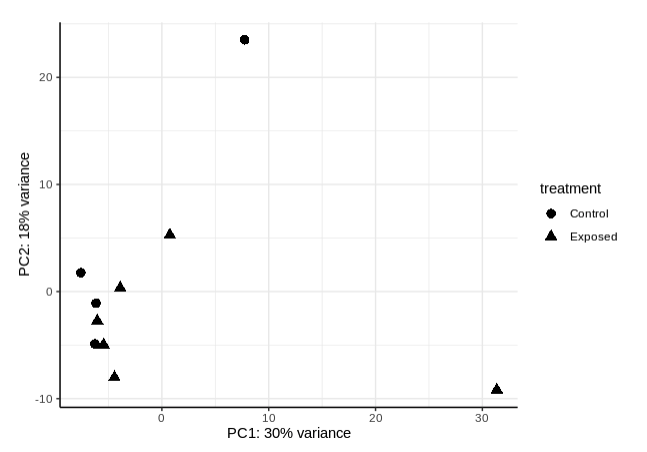
PCA by treatment:
pickSoftThrshold plot:
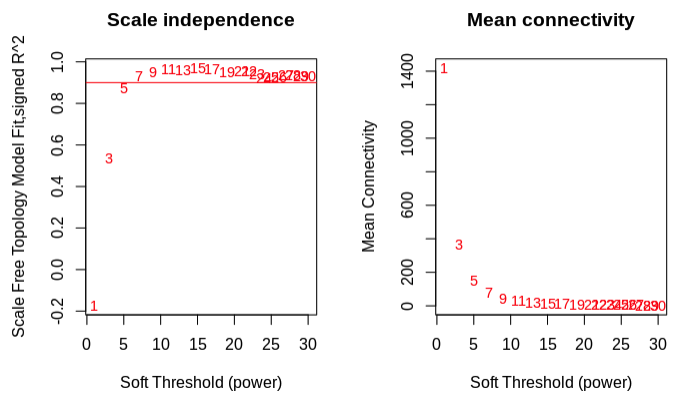
- Chose 5
Distance matrix from TOM dissimilarity and plot genet tree:
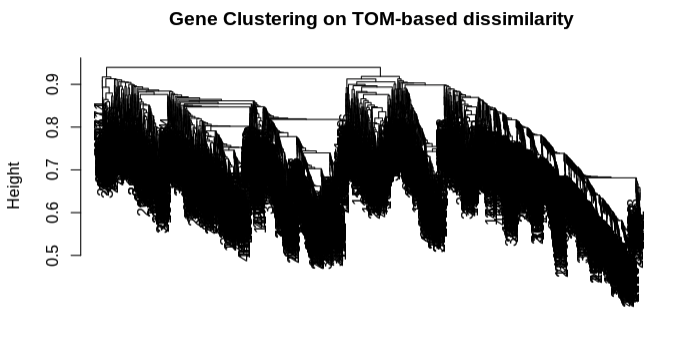
- Clustering looks good
Identify modules of lncRNAs with similar expression patterns:
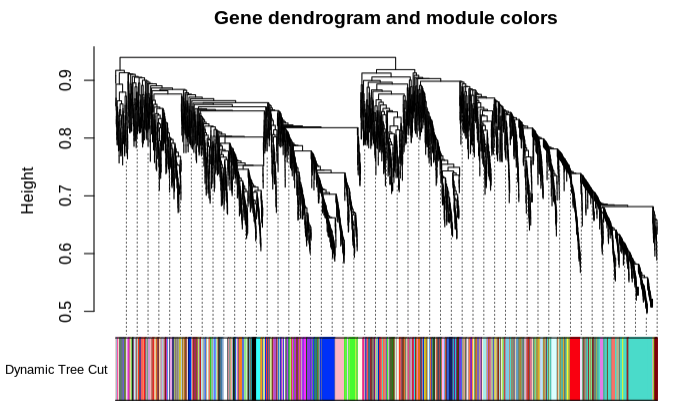
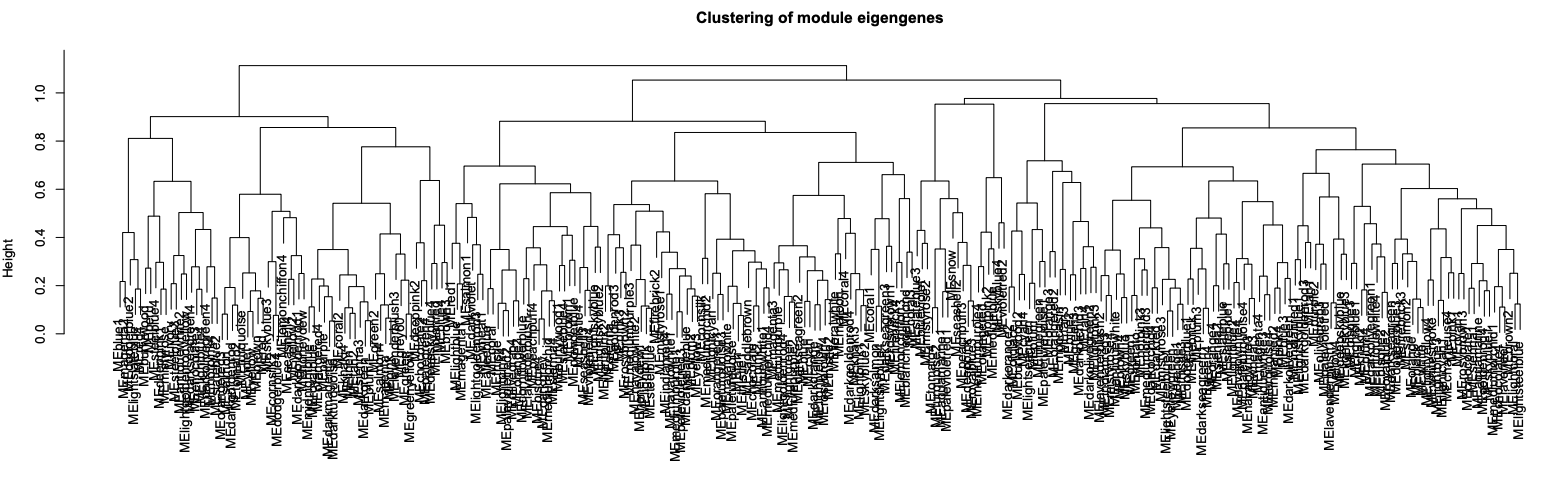
Module similarity based on eigengene value:
Merge modules with >85% eigengene similarity and plot:
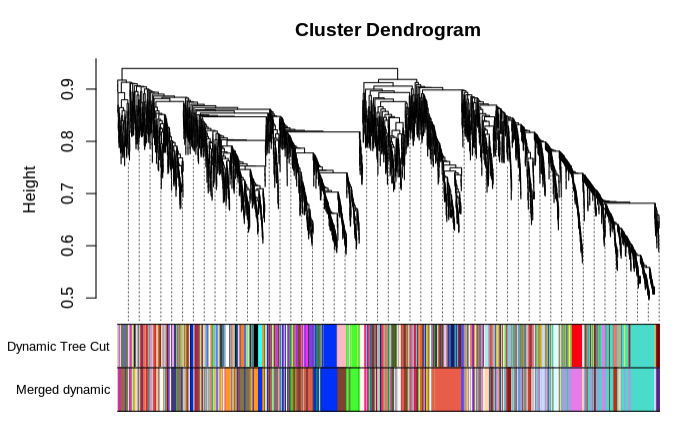
- 185 modules total
| Color Name | Count |
|---|---|
| aliceblue | 20 |
| antiquewhite | 7 |
| antiquewhite1 | 18 |
| antiquewhite2 | 12 |
| antiquewhite3 | 11 |
| antiquewhite4 | 15 |
| aquamarine | 5 |
| aquamarine1 | 16 |
| bisque2 | 12 |
| bisque3 | 6 |
| bisque4 | 75 |
| blanchedalmond | 7 |
| blue | 265 |
| blue1 | 8 |
| blue2 | 13 |
| blue3 | 9 |
| blue4 | 11 |
| blueviolet | 11 |
| brown1 | 9 |
| brown2 | 13 |
| brown3 | 8 |
| brown4 | 48 |
| burlywood | 7 |
| burlywood1 | 6 |
| burlywood2 | 24 |
| chartreuse4 | 5 |
| chocolate | 13 |
| chocolate1 | 14 |
| chocolate2 | 7 |
| chocolate3 | 19 |
| coral1 | 15 |
| coral2 | 336 |
| coral3 | 12 |
| coral4 | 10 |
| cornflowerblue | 8 |
| cornsilk | 7 |
| cornsilk2 | 13 |
| cyan1 | 5 |
| cyan4 | 5 |
| darkgoldenrod1 | 6 |
| darkgoldenrod3 | 7 |
| darkgoldenrod4 | 8 |
| darkolivegreen | 26 |
| darkolivegreen1 | 9 |
| darkolivegreen2 | 11 |
| darkolivegreen4 | 13 |
| darkorange2 | 25 |
| darkorchid3 | 5 |
| darkorchid4 | 39 |
| darksalmon | 15 |
| darkseagreen | 7 |
| darkseagreen1 | 18 |
| darkseagreen2 | 10 |
| darkseagreen3 | 12 |
| darkseagreen4 | 15 |
| darkslateblue | 72 |
| darkviolet | 13 |
| deeppink | 20 |
| deeppink1 | 19 |
| deeppink2 | 8 |
| deepskyblue | 7 |
| deepskyblue4 | 6 |
| dodgerblue1 | 5 |
| dodgerblue2 | 5 |
| dodgerblue3 | 6 |
| firebrick | 8 |
| firebrick2 | 9 |
| firebrick3 | 11 |
| firebrick4 | 13 |
| floralwhite | 19 |
| goldenrod3 | 5 |
| green | 145 |
| green1 | 15 |
| green2 | 7 |
| green3 | 8 |
| honeydew | 12 |
| honeydew1 | 74 |
| hotpink2 | 5 |
| hotpink3 | 5 |
| hotpink4 | 13 |
| indianred | 7 |
| indianred1 | 8 |
| indianred2 | 20 |
| indianred3 | 35 |
| ivory | 19 |
| khaki2 | 16 |
| khaki3 | 27 |
| lavenderblush | 8 |
| lavenderblush1 | 24 |
| lavenderblush2 | 12 |
| lavenderblush3 | 15 |
| lemonchiffon3 | 5 |
| lemonchiffon4 | 5 |
| lightblue | 6 |
| lightblue1 | 7 |
| lightblue2 | 8 |
| lightblue3 | 28 |
| lightcoral | 26 |
Generate a complex heatmap of module-trait relationships.
-
heatmap found here
-
Examine mean module eigengene values for modules of interest (those that are significant from the complex heatmap)
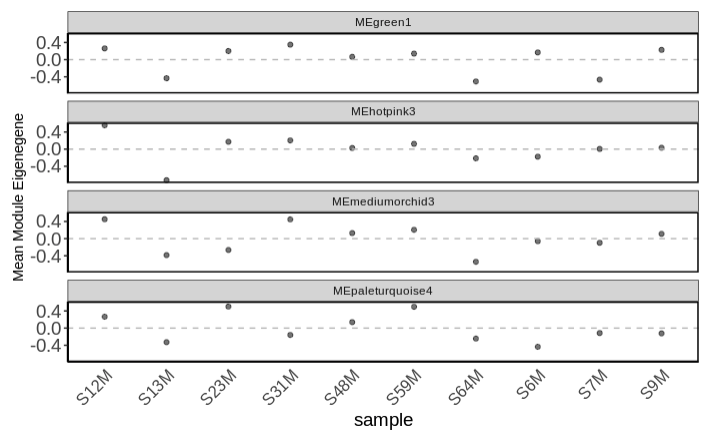
-
Plot mean eigen values by treatment.
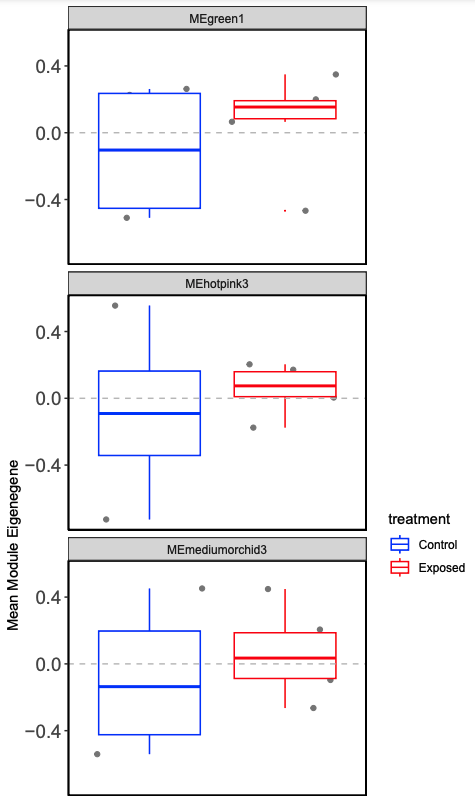
Fewer modules than females (185 compare to 238). Similar number of significantly different modules by treatment.