Started with a couple smaller datasets included in the umbrella of RNAcentral, but got almost no hits so scaled up. Blasted the entire RNAcentral database (over 3 million sequences) with the Crassostrea virginica lncRNA FASTA file.
Key Points:
- This isn’t really an annotation, just a database comparison to see if any of the lncRNAs are better described in other speacies
- Was able to get a hit for every lncRNA transcript, but this isn’t surprising since the database includes uncharacterized RNAs from C. virginica in the Ensembl Metazoa database.
- Not sure trying to annotate this way does anything meaningful for us since the count matrix already uses the IDs associated with these catalogged but uncharacterized RNAs.
Summary stats of blast run:
- All 4750 lncRNA transcripts obtained 1 or greater hits
- Results table (hits are represented by RNAcentral IDs)
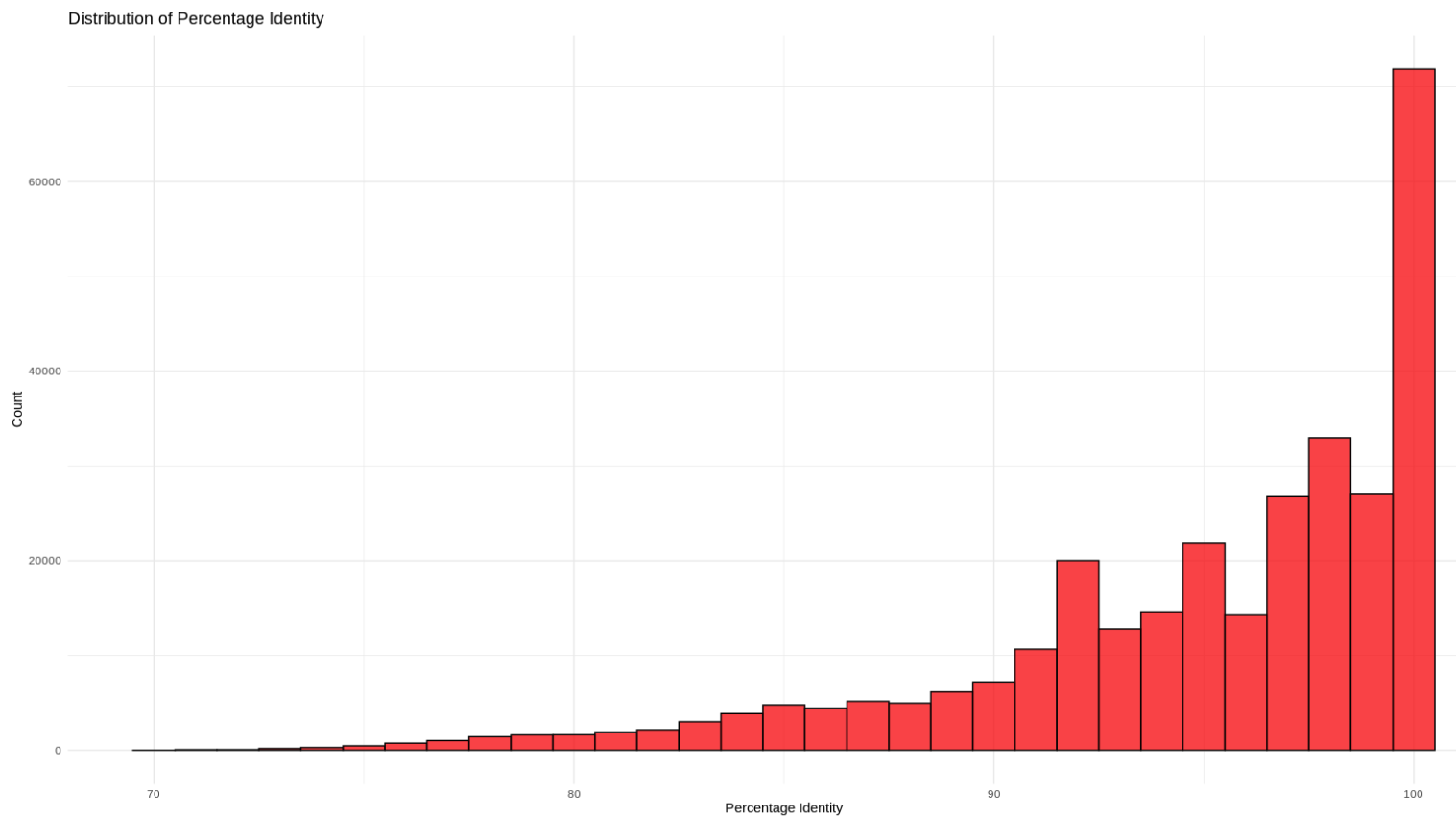
Distribution of Percentage Identity
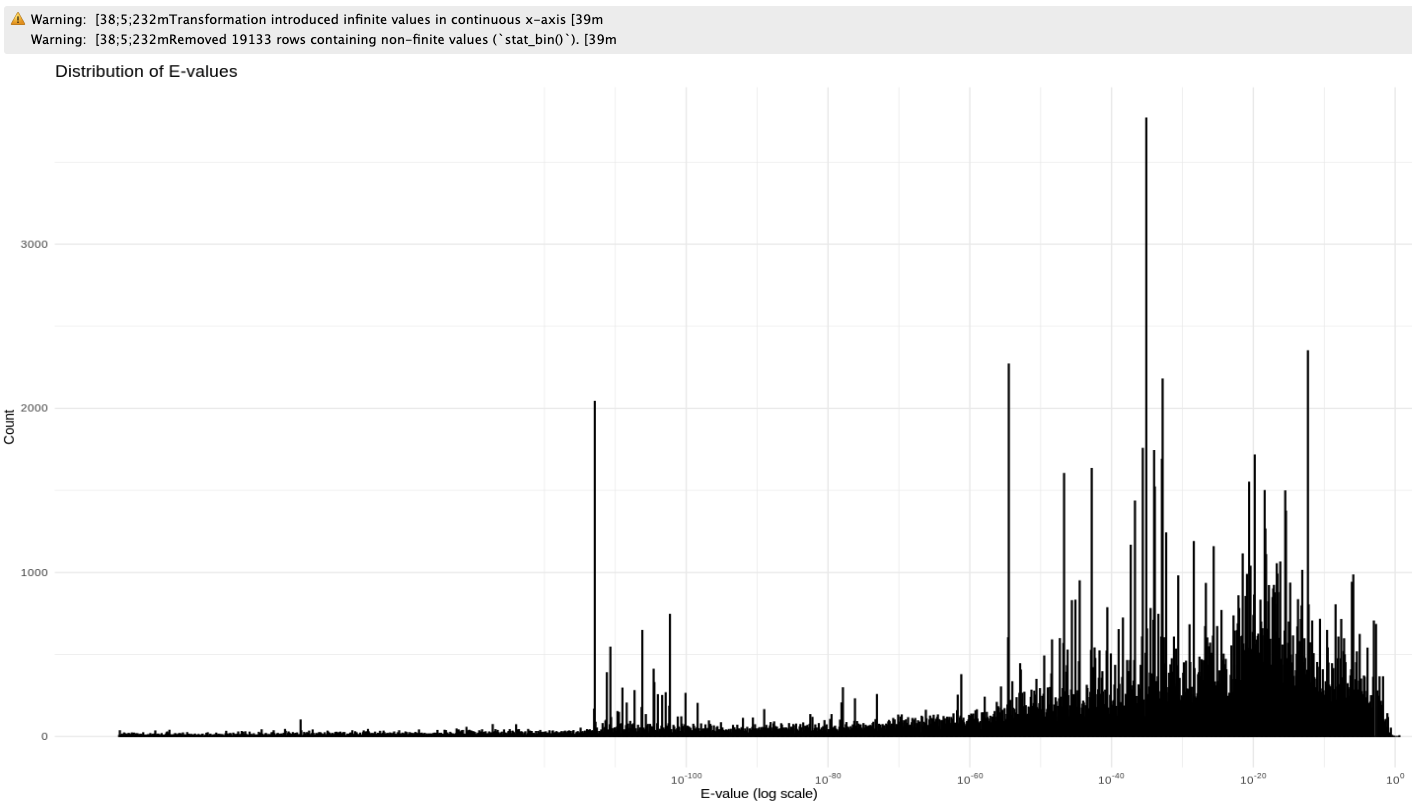
E-value Distribution