Using the Google Earth Engine platform to load, process, analyze, and visualize remote sensing data
TL;DR Notice: Start at the “Products and Interface” section if you aren’t interested in background
Prior to joining the Roberts Lab this Fall, I worked for NASA Earth Applied Sciences’ DEVELOP and ARSET programs. During this time, I became familiar a variety of environmental remote sensing techniques and platforms. One of the easiest ways to work with remote sensing data is Google Earth Engine (GEE). GEE is a cloud-based raster computing platforms that uses a JavaScript API to manipulate remote sensing data. A variety of satellite images and data products are hosted through GEE (including Landsat and Sentinel imagery). This negates data storage issues and local machine computing limits. If you would like to learn more about GEE in general, please refer to this training I gave in June.
 The Google Earth Engine interface. Access to GEE is free, so you can sign up for an account at no cost. Free access is guaranteed to scientists, students, and natural resource managers for the foreseeable future.
The Google Earth Engine interface. Access to GEE is free, so you can sign up for an account at no cost. Free access is guaranteed to scientists, students, and natural resource managers for the foreseeable future.
Aquatic environmental parameter assessment and using the Optical Reef and Coastal Area Assessment (ORCAA) Tool
For the purposes of coral reef ecology, satellite remote sensing has the potential to expand spatial and temporal coverage of parameters such as chlorophyll, turbidity, SST, and CDOM. Public satellite technology made available by government agencies (namely NASA and ESA) can provide data at a 10m-30m resolution. Temporal resolution varies between sensors, but using data from more than one satellite sensor can provide multiple images per month.
During my time at NASA DEVELOP a GEE tool was developed for remote assessment of various parameters related to reef and coastal water quality using imagery from the NASA Landsat 8 OLI sensor and the ESA Sentinel-2 MSI sensor. The tool implements a graphical user interface (GUI) giving the user the ability to call a time series of imagery over a specified period and spatial extent, providing a ready made platform to alter and specialize for the application of water quality algorithms according to the needs of research goals.
Products and Interface
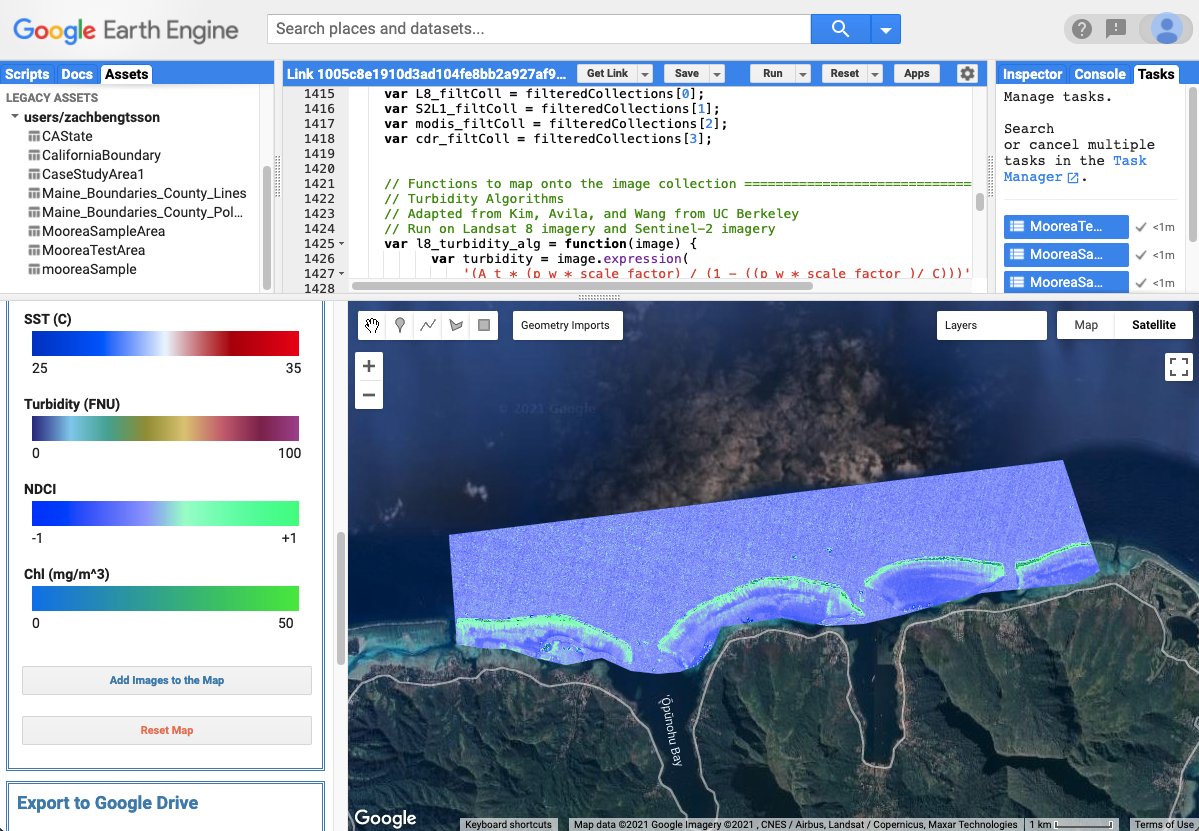 ORCAA Interface displaying NDCI
ORCAA Interface displaying NDCI
Technical information in this section was originally prepared by the creators of the ORCAA tool: Hayley Pippin, Arbyn Olarte, Roxana Pilot, and Vanessa Valenti
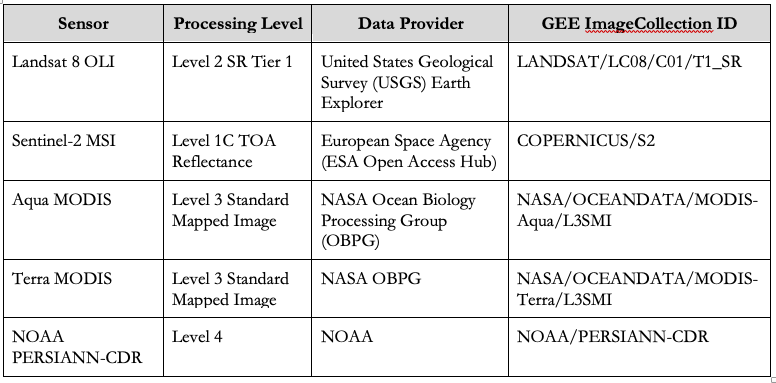
Data inputs from common NASA and ESA sensors were used via the GEE internal data catalog:
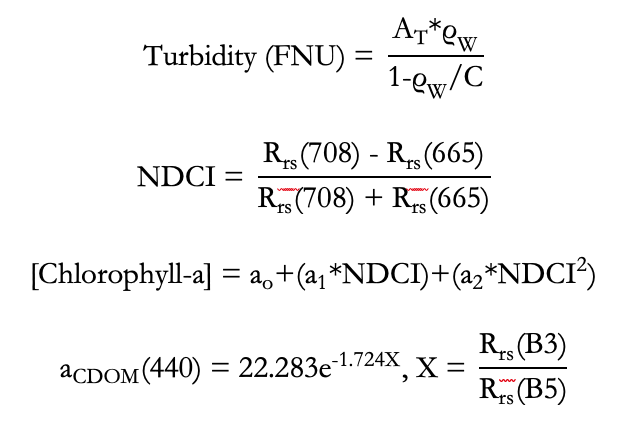
Parameters include Turbidity, Normalized Difference Chlorophyll Index (NDCI) and Chlorophyll-a, Dissolved Organic Matter (CDOM), and Sea Surface Temperature (SST). Links are to publications which established the following equations for calculation of these parameters from surface reflectance bands:
Interface allows you to select dates of interest and upload your own spatial boundaries:

I chose an aggregate of data from Jan - Nov in 2019 in this example over a polygon drawn in GEE (below).
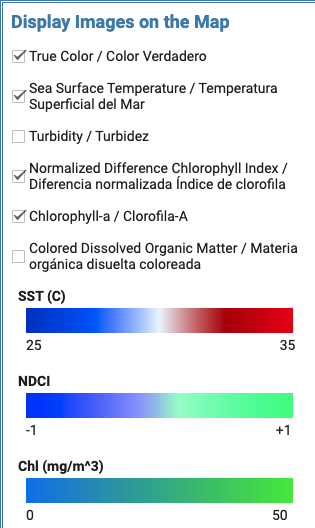
Check parameters you want to retrieve, calculate, and display. Relevant colorscales will populate:
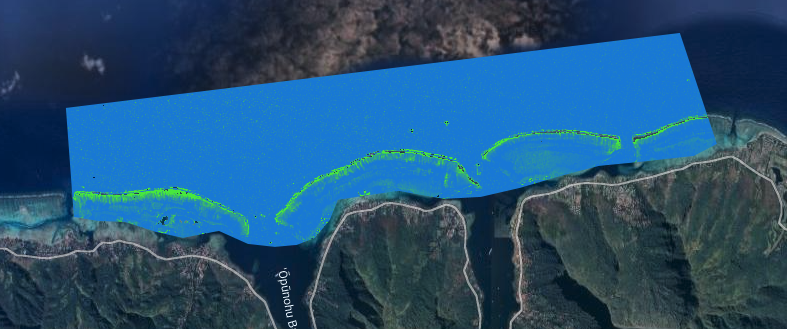
After you run, layers are made available in the viewer. Including the highest quality true color satellite image available in your date range:

Chlorophyll-a layer:

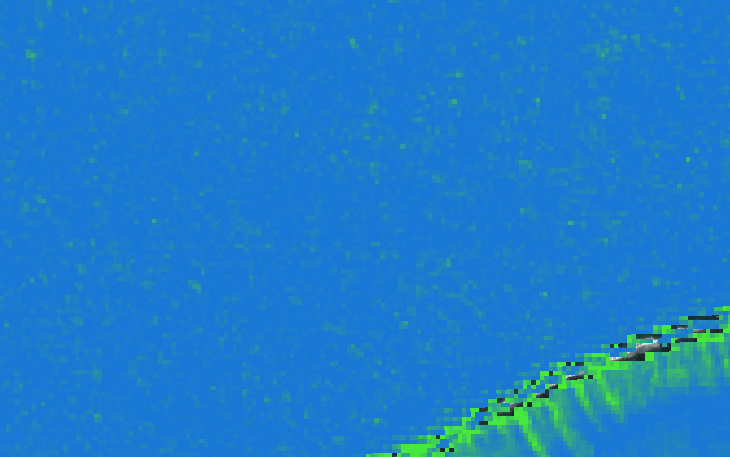
Zoom for pixel by pixel view:
Time series chart generator allows you to display data over the course of your data range, values are an average of all pixels within your spatial boundary:
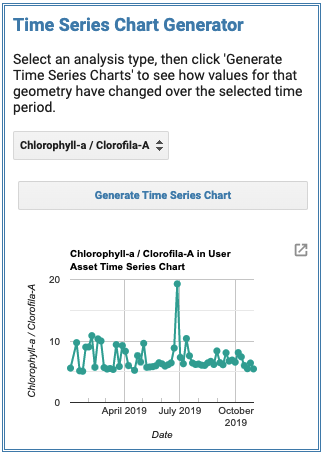
Imagery, data layers, time series graphs, and point data are all exportable in TIFF, JPEG, and CSV format. Updates under way by the NASA DEVELOP program are implementing new cloud filtering data and improved surface reflectance values. This format of data retrieval, manipulation, and display is particularly helpful in deciding which data products may be most useful in research and allow for the quick processing of satellite imagery.
Take a look at the ORCAA repository for further information or use the link here (you’ll need a GEE account) to test it out yourself.